R & D life with iPython notebook
This is a memo for working while staying at home or Starbucks using iPython notebook, which is a Python web interface. I will update it when I feel like it.
Allow access to notebooks on remote servers
You can start your R & D with Starbucks at any time by launching your notebook on your workstation at work. Please refer to here for the introduction. `c.IPKernelApp.pylab ='inline'` is especially important, so that what you draw with matplotlib can be displayed on your notebook.
- If you want to use free Wi-Fi with Starbucks, here
- The store where you can drink delicious Starbucks coffee is here
Run the program
Create a new notebook for each test so you don't get confused. The following Cell magic is useful for debugging.
`% time` Measure the time
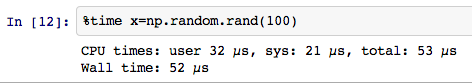
`` `% prun``` profiling

`% debug` Start the debugger
After the program stops with an error, run `` `% debug``` on the appropriate Cell to start ipdb
Calculate in parallel
There are several methods. This area is also helpful I will.
1 multiprocessing.Pool
Refer to this area
from multiprocessing import Pool
p = Pool(n_cores)
p.map(func, [arg list])
p.close()
You can calculate in parallel like this. It should be noted that in the middle of running parallel computing If you press the suspend button or restart button here, the worker will usually become a zombie. In that case, I try to kill everything with `` `pkill Python``` etc. and then start up the notebook again.
2 IPython.parallel
Since iPython itself has a framework for parallel computing, it is one way to use it. First, launch iPython notebook, select the Clusters tab from the dashboard that appears first, and then
 Enter the desired number of workers in # of engines here and start.
In the program, write as follows.
Enter the desired number of workers in # of engines here and start.
In the program, write as follows.
from IPython.parallel import Client
def test(x):
u'''
Functions calculated in parallel
'''
return x **2
cli = Client()
dv = cli[:]
x = dv.map(test, range(100))
result = x.get()
3 Run the program itself in parallel
If you want to run the same function multiple times in parallel while changing the arguments, methods 1 and 2 are effective, but if you want to run a large program in parallel, it is easier to perform background processing with a shell script and parallelize it. In other words, let's execute a shell script that writes multiple `python program.py arg1 arg2 ... &`.
You can also run a bash script on a notebook Cell by using the `%% bash` command, you said, "I was talking about iPython notebook so far, but it's a shell after all!" I will. For example

If you change this a little,
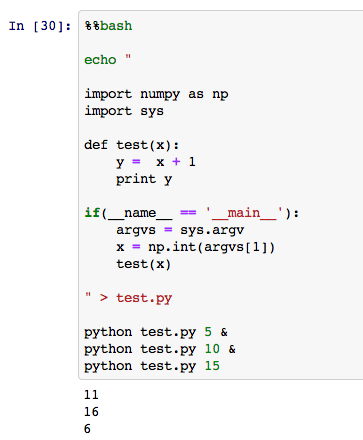
You can easily (?) Parallelize like this. If you want to use command line arguments in Python, create a if (__ name__ =='__main__'): `` `block and read it from` `sys.argv.
Allows you to hit SSH from Chrome
If you have a small program, you can write it on iPython notebook, but when the program gets messy, it is better to make it a .py file, so you will inevitably need to dive into the workstation with SSH. .. I'm using Secure Shell ( link ) ..
- It is convenient to enable terminal multiplexing (for example, tmux ) so that work can be interrupted.
- gedi-vim or <a href="https://github.com/klen/python -mode "target =" _ blank "> python -mode is useful
- Both should have both vim / emacs versions I think there are many people who recommend + gedi, but I am using python-mode because it is an amanojaku. It is convenient to rename and complete variables, and to fix parts that do not conform to PEP8.
Manage python packages
I use Canopy . OpenCV is also convenient because it can be inserted with one click. You can use the enpkg command to manage packages from the terminal (<a href="https://support.enthought.com/entries/22415022-Using-enpkg-to-update-Canopy-EPD" -packages "target =" _ blank "> link ). Mac users may be addicted to keychain issues, so <a href="http://stackoverflow.com/questions/14719731/keychain-issue-when-trying-to-set-up-enthought-enpkg-on -check mac-os-x "target =" _ blank "> here as well.
Recommended Posts