Pandas User Guide "Table Formatting and PivotTables" (Official Document Japanese Translation)
This article is part of the official Pandas documentation after machine translation of the User Guide --Reshaping and pivot tables (https://pandas.pydata.org/pandas-docs/stable/user_guide/reshaping.html). It is a modification of an unnatural sentence.
If you have any mistranslations, alternative translations, questions, etc., please use the comments section or edit request.
Table shaping and pivot table
Formatting by pivoting a DataFrame object
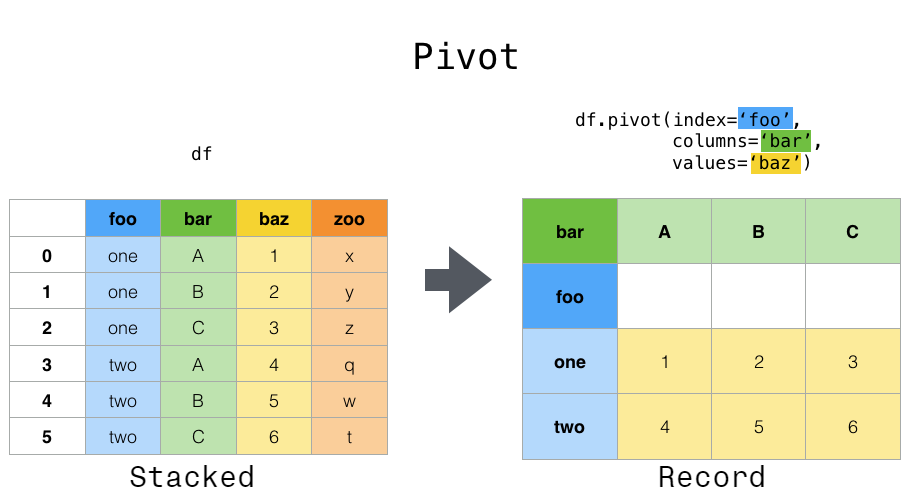
Data is often stored in so-called "stacks" or "records".
In [1]: df
Out[1]:
date variable value
0 2000-01-03 A 0.469112
1 2000-01-04 A -0.282863
2 2000-01-05 A -1.509059
3 2000-01-03 B -1.135632
4 2000-01-04 B 1.212112
5 2000-01-05 B -0.173215
6 2000-01-03 C 0.119209
7 2000-01-04 C -1.044236
8 2000-01-05 C -0.861849
9 2000-01-03 D -2.104569
10 2000-01-04 D -0.494929
11 2000-01-05 D 1.071804
By the way, how to create the above DataFrame is as follows.
import pandas._testing as tm
def unpivot(frame):
N, K = frame.shape
data = {'value': frame.to_numpy().ravel('F'),
'variable': np.asarray(frame.columns).repeat(N),
'date': np.tile(np.asarray(frame.index), K)}
return pd.DataFrame(data, columns=['date', 'variable', 'value'])
df = unpivot(tm.makeTimeDataFrame(3))
To select all rows where variable is ʻA`:
In [2]: df[df['variable'] == 'A']
Out[2]:
date variable value
0 2000-01-03 A 0.469112
1 2000-01-04 A -0.282863
2 2000-01-05 A -1.509059
On the other hand, suppose you want to use these variables to perform time series operations. In this case, it would be better to identify the individual observations by the unique variable columns and the date ʻindex. To reshape your data into this format, [DataFrame.pivot ()](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.pivot.html#pandas Use the .DataFrame.pivot) method (top-level function [pivot ()`](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.pivot.html#pandas) .pivot) is also implemented).
In [3]: df.pivot(index='date', columns='variable', values='value')
Out[3]:
variable A B C D
date
2000-01-03 0.469112 -1.135632 0.119209 -2.104569
2000-01-04 -0.282863 1.212112 -1.044236 -0.494929
2000-01-05 -1.509059 -0.173215 -0.861849 1.071804
If the values argument is omitted and the input DataFrame has multiple columns of values that are not given to pivot as columns or indexes, the resulting" pivot " DataFrame will have the highest level of each value column. Has a Hierarchical Column (https://qiita.com/nkay/items/ Multi-Index Advanced Index) that indicates.
In [4]: df['value2'] = df['value'] * 2
In [5]: pivoted = df.pivot(index='date', columns='variable')
In [6]: pivoted
Out[6]:
value ... value2
variable A B C ... B C D
date ...
2000-01-03 0.469112 -1.135632 0.119209 ... -2.271265 0.238417 -4.209138
2000-01-04 -0.282863 1.212112 -1.044236 ... 2.424224 -2.088472 -0.989859
2000-01-05 -1.509059 -0.173215 -0.861849 ... -0.346429 -1.723698 2.143608
[3 rows x 8 columns]
You can then select a subset from the pivoted DataFrame.
In [7]: pivoted['value2']
Out[7]:
variable A B C D
date
2000-01-03 0.938225 -2.271265 0.238417 -4.209138
2000-01-04 -0.565727 2.424224 -2.088472 -0.989859
2000-01-05 -3.018117 -0.346429 -1.723698 2.143608
Note that if the data is isomorphic, it returns a view of the underlying data.
: ballot_box_with_check: ** Note ** If the index / column pair is not unique,
pivot ()will Raises the errorValueError: Index contains duplicate entries, cannot reshape. In this case, consider usingpivot_table ()Please give me. This is a generalization of pivots that can handle duplicate values for a single index / column pair.
Shape change by stacking and unstacking
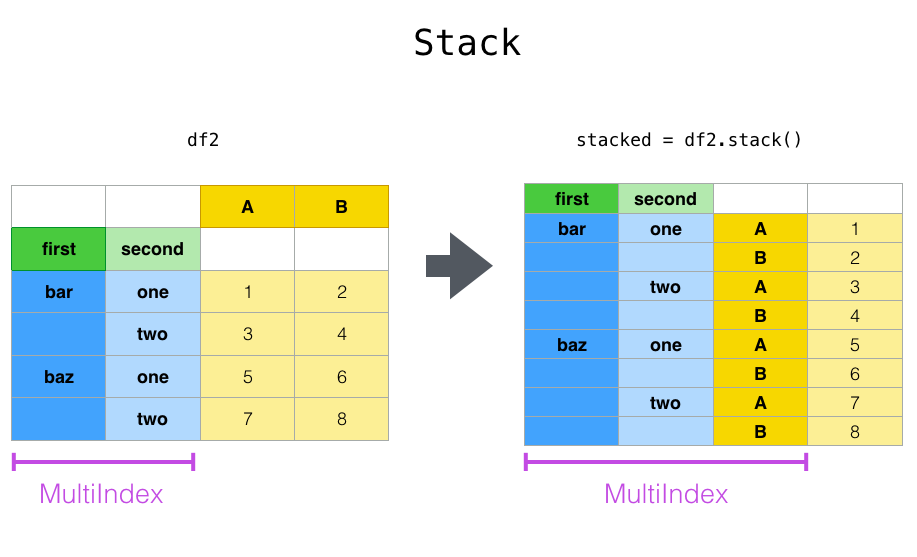
pivot () A series closely related to the method In the method of, stack () available in Series and DataFrame # pandas.DataFrame.stack) and [ʻunstack () ](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.unstack.html#pandas.DataFrame.unstack) There is a method. These methods are designed to work with the MultiIndex` object (see the chapter on Hierarchical Indexes (https://qiita.com/nkay/items/Multiindex Advanced Indexes)). The basic functions of these methods are as follows:
--stack:" Pivots "the level of the (possibly hierarchical) column label and returns a new DataFrame with that row label at the innermost level of the index.
--ʻUnstack: (reverse stacking) (probably hierarchical) row index level "pivot" to column axis to generate reconstructed DataFrame` with innermost level column label I will.
It will be easier to understand if you look at an actual example. It deals with the same dataset that we saw in the Hierarchical Index chapter.
In [8]: tuples = list(zip(*[['bar', 'bar', 'baz', 'baz',
...: 'foo', 'foo', 'qux', 'qux'],
...: ['one', 'two', 'one', 'two',
...: 'one', 'two', 'one', 'two']]))
...:
In [9]: index = pd.MultiIndex.from_tuples(tuples, names=['first', 'second'])
In [10]: df = pd.DataFrame(np.random.randn(8, 2), index=index, columns=['A', 'B'])
In [11]: df2 = df[:4]
In [12]: df2
Out[12]:
A B
first second
bar one 0.721555 -0.706771
two -1.039575 0.271860
baz one -0.424972 0.567020
two 0.276232 -1.087401
The stack function" compresses "the level of a column in DataFrame to produce one of the following:
--Series: For simple column indexes.
--DataFrame: If the column has MultiIndex.
If the column has MultiIndex, you can choose the level to stack. The stacked level will be the lowest level of the new column MultiIndex.
In [13]: stacked = df2.stack()
In [14]: stacked
Out[14]:
first second
bar one A 0.721555
B -0.706771
two A -1.039575
B 0.271860
baz one A -0.424972
B 0.567020
two A 0.276232
B -1.087401
dtype: float64
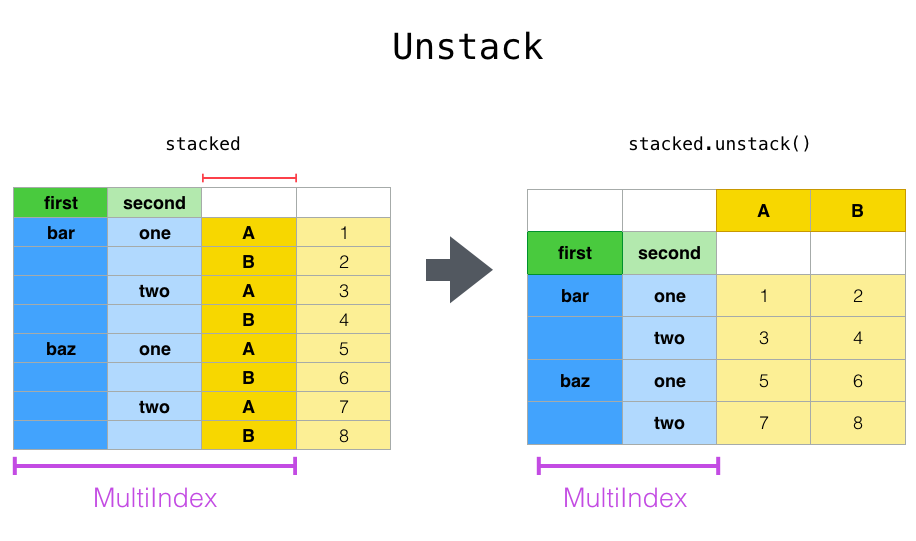
For a "stacked" DataFrame or Series (that is, ʻindex is MultiIndex), use ʻunstack to perform the opposite of stack. The default is to unstack the ** lowest level **.
In [15]: stacked.unstack()
Out[15]:
A B
first second
bar one 0.721555 -0.706771
two -1.039575 0.271860
baz one -0.424972 0.567020
two 0.276232 -1.087401
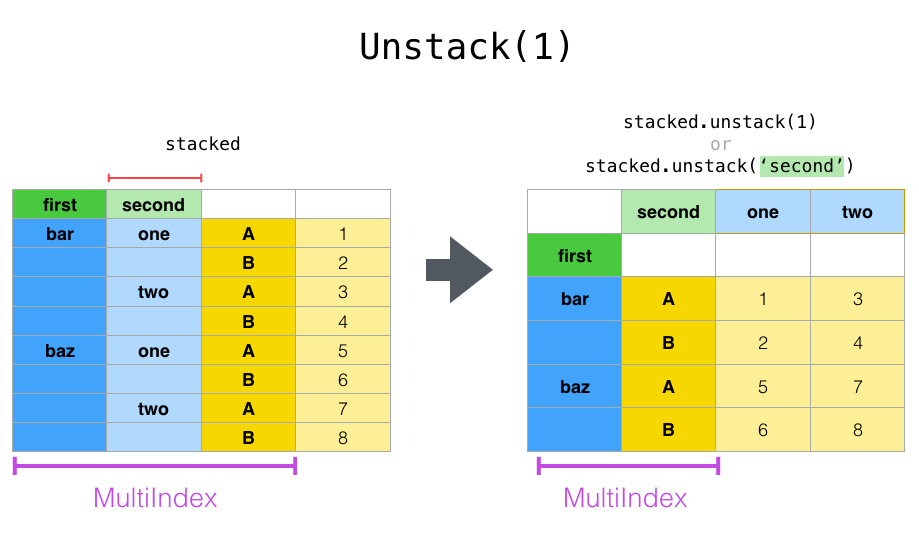
In [16]: stacked.unstack(1)
Out[16]:
second one two
first
bar A 0.721555 -1.039575
B -0.706771 0.271860
baz A -0.424972 0.276232
B 0.567020 -1.087401
In [17]: stacked.unstack(0)
Out[17]:
first bar baz
second
one A 0.721555 -0.424972
B -0.706771 0.567020
two A -1.039575 0.276232
B 0.271860 -1.087401
If the index has a name, you can use the level name instead of specifying the level number.
In [18]: stacked.unstack('second')
Out[18]:
second one two
first
bar A 0.721555 -1.039575
B -0.706771 0.271860
baz A -0.424972 0.276232
B 0.567020 -1.087401
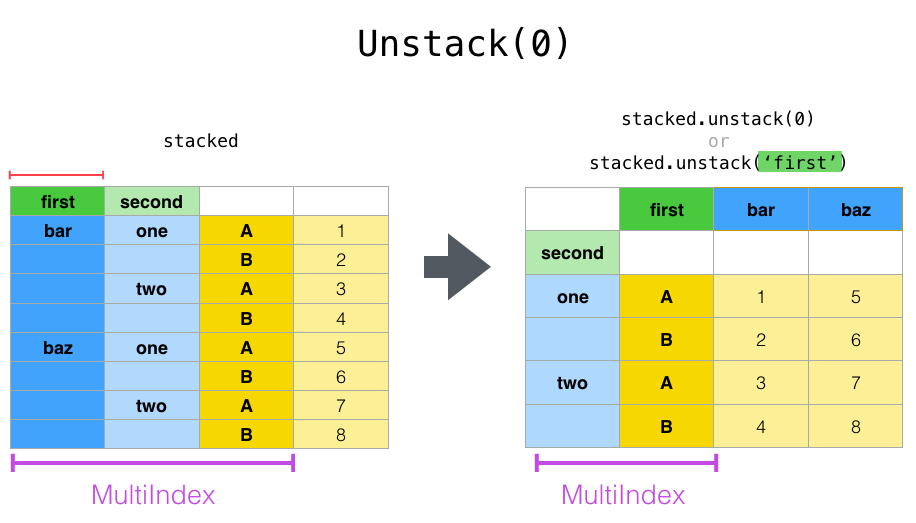
Note that the stack and ʻunstackmethods implicitly sort the associated index levels. Therefore,stack and then ʻunstack (or vice versa) will return a ** sorted ** copy of the original DataFrame or Series.
In [19]: index = pd.MultiIndex.from_product([[2, 1], ['a', 'b']])
In [20]: df = pd.DataFrame(np.random.randn(4), index=index, columns=['A'])
In [21]: df
Out[21]:
A
2 a -0.370647
b -1.157892
1 a -1.344312
b 0.844885
In [22]: all(df.unstack().stack() == df.sort_index())
Out[22]: True
The above code will throw a TypeError if the call to sort_index is removed.
Multiple levels
You can also stack or unstack multiple levels at once by passing a list of levels. In that case, the end result is the same as if each level in the list was processed individually.
In [23]: columns = pd.MultiIndex.from_tuples([
....: ('A', 'cat', 'long'), ('B', 'cat', 'long'),
....: ('A', 'dog', 'short'), ('B', 'dog', 'short')],
....: names=['exp', 'animal', 'hair_length']
....: )
....:
In [24]: df = pd.DataFrame(np.random.randn(4, 4), columns=columns)
In [25]: df
Out[25]:
exp A B A B
animal cat cat dog dog
hair_length long long short short
0 1.075770 -0.109050 1.643563 -1.469388
1 0.357021 -0.674600 -1.776904 -0.968914
2 -1.294524 0.413738 0.276662 -0.472035
3 -0.013960 -0.362543 -0.006154 -0.923061
In [26]: df.stack(level=['animal', 'hair_length'])
Out[26]:
exp A B
animal hair_length
0 cat long 1.075770 -0.109050
dog short 1.643563 -1.469388
1 cat long 0.357021 -0.674600
dog short -1.776904 -0.968914
2 cat long -1.294524 0.413738
dog short 0.276662 -0.472035
3 cat long -0.013960 -0.362543
dog short -0.006154 -0.923061
The list of levels can contain either level names or level numbers (although the two cannot be mixed).
# df.stack(level=['animal', 'hair_length'])
#This code is equal to
In [27]: df.stack(level=[1, 2])
Out[27]:
exp A B
animal hair_length
0 cat long 1.075770 -0.109050
dog short 1.643563 -1.469388
1 cat long 0.357021 -0.674600
dog short -1.776904 -0.968914
2 cat long -1.294524 0.413738
dog short 0.276662 -0.472035
3 cat long -0.013960 -0.362543
dog short -0.006154 -0.923061
Missing data
These functions are also flexible in handling missing data and will work even if each subgroup in the hierarchical index does not have the same set of labels. You can also handle unsorted indexes (of course, you can sort by calling sort_index). Here is a more complex example:
In [28]: columns = pd.MultiIndex.from_tuples([('A', 'cat'), ('B', 'dog'),
....: ('B', 'cat'), ('A', 'dog')],
....: names=['exp', 'animal'])
....:
In [29]: index = pd.MultiIndex.from_product([('bar', 'baz', 'foo', 'qux'),
....: ('one', 'two')],
....: names=['first', 'second'])
....:
In [30]: df = pd.DataFrame(np.random.randn(8, 4), index=index, columns=columns)
In [31]: df2 = df.iloc[[0, 1, 2, 4, 5, 7]]
In [32]: df2
Out[32]:
exp A B A
animal cat dog cat dog
first second
bar one 0.895717 0.805244 -1.206412 2.565646
two 1.431256 1.340309 -1.170299 -0.226169
baz one 0.410835 0.813850 0.132003 -0.827317
foo one -1.413681 1.607920 1.024180 0.569605
two 0.875906 -2.211372 0.974466 -2.006747
qux two -1.226825 0.769804 -1.281247 -0.727707
As mentioned earlier, you can call stack with the level argument to choose which level in the column to stack.
In [33]: df2.stack('exp')
Out[33]:
animal cat dog
first second exp
bar one A 0.895717 2.565646
B -1.206412 0.805244
two A 1.431256 -0.226169
B -1.170299 1.340309
baz one A 0.410835 -0.827317
B 0.132003 0.813850
foo one A -1.413681 0.569605
B 1.024180 1.607920
two A 0.875906 -2.006747
B 0.974466 -2.211372
qux two A -1.226825 -0.727707
B -1.281247 0.769804
In [34]: df2.stack('animal')
Out[34]:
exp A B
first second animal
bar one cat 0.895717 -1.206412
dog 2.565646 0.805244
two cat 1.431256 -1.170299
dog -0.226169 1.340309
baz one cat 0.410835 0.132003
dog -0.827317 0.813850
foo one cat -1.413681 1.024180
dog 0.569605 1.607920
two cat 0.875906 0.974466
dog -2.006747 -2.211372
qux two cat -1.226825 -1.281247
dog -0.727707 0.769804
If the subgroups do not have the same set of labels, unstacking can cause missing values. By default, missing values are replaced with the default fill-in-the-blank values for that data type (such as NaN for floats, NaT for datetime-like). For integer types, the data is converted to float by default and the missing value is set to NaN.
In [35]: df3 = df.iloc[[0, 1, 4, 7], [1, 2]]
In [36]: df3
Out[36]:
exp B
animal dog cat
first second
bar one 0.805244 -1.206412
two 1.340309 -1.170299
foo one 1.607920 1.024180
qux two 0.769804 -1.281247
In [37]: df3.unstack()
Out[37]:
exp B
animal dog cat
second one two one two
first
bar 0.805244 1.340309 -1.206412 -1.170299
foo 1.607920 NaN 1.024180 NaN
qux NaN 0.769804 NaN -1.281247
ʻUnstack can also take an optional fill_value` argument that specifies the value of the missing data.
In [38]: df3.unstack(fill_value=-1e9)
Out[38]:
exp B
animal dog cat
second one two one two
first
bar 8.052440e-01 1.340309e+00 -1.206412e+00 -1.170299e+00
foo 1.607920e+00 -1.000000e+09 1.024180e+00 -1.000000e+09
qux -1.000000e+09 7.698036e-01 -1.000000e+09 -1.281247e+00
For MultiIndex
Even when the column is MultiIndex, it can be unstacked without any problems.
In [39]: df[:3].unstack(0)
Out[39]:
exp A B ... A
animal cat dog ... cat dog
first bar baz bar ... baz bar baz
second ...
one 0.895717 0.410835 0.805244 ... 0.132003 2.565646 -0.827317
two 1.431256 NaN 1.340309 ... NaN -0.226169 NaN
[2 rows x 8 columns]
In [40]: df2.unstack(1)
Out[40]:
exp A B ... A
animal cat dog ... cat dog
second one two one ... two one two
first ...
bar 0.895717 1.431256 0.805244 ... -1.170299 2.565646 -0.226169
baz 0.410835 NaN 0.813850 ... NaN -0.827317 NaN
foo -1.413681 0.875906 1.607920 ... 0.974466 0.569605 -2.006747
qux NaN -1.226825 NaN ... -1.281247 NaN -0.727707
[4 rows x 8 columns]
Plastic surgery by melt
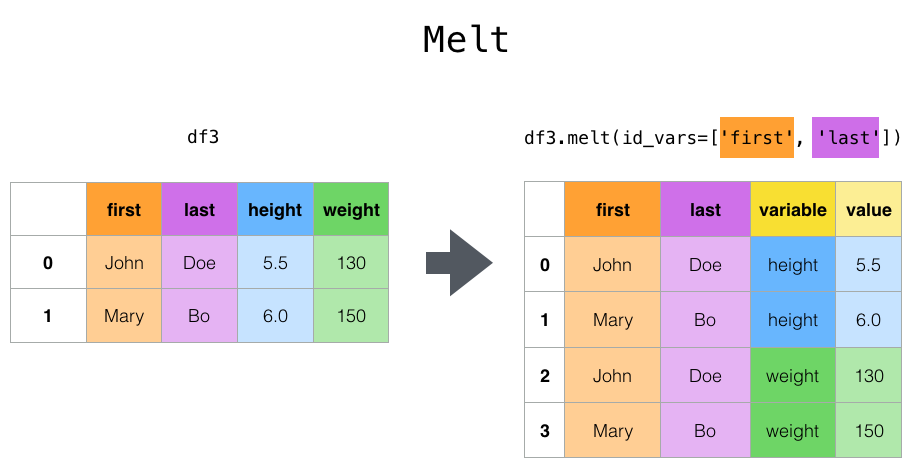
Top-level melt () functions and their corresponding DataFrame. melt () has one or more DataFrame Converts to an "unpivoted" state where the column of is regarded as a * identification variable * and all other columns are regarded as * measured variables *, leaving only two non-identifying columns, "variable" and "value". Useful for. The names of the remaining columns can be customized by specifying the var_name and value_name parameters.
For example
In [41]: cheese = pd.DataFrame({'first': ['John', 'Mary'],
....: 'last': ['Doe', 'Bo'],
....: 'height': [5.5, 6.0],
....: 'weight': [130, 150]})
....:
In [42]: cheese
Out[42]:
first last height weight
0 John Doe 5.5 130
1 Mary Bo 6.0 150
In [43]: cheese.melt(id_vars=['first', 'last'])
Out[43]:
first last variable value
0 John Doe height 5.5
1 Mary Bo height 6.0
2 John Doe weight 130.0
3 Mary Bo weight 150.0
In [44]: cheese.melt(id_vars=['first', 'last'], var_name='quantity')
Out[44]:
first last quantity value
0 John Doe height 5.5
1 Mary Bo height 6.0
2 John Doe weight 130.0
3 Mary Bo weight 150.0
As another conversion method, wide_to_long () is useful for panel data. It is a function. Less flexible than melt (), but more user It's friendly.
In [45]: dft = pd.DataFrame({"A1970": {0: "a", 1: "b", 2: "c"},
....: "A1980": {0: "d", 1: "e", 2: "f"},
....: "B1970": {0: 2.5, 1: 1.2, 2: .7},
....: "B1980": {0: 3.2, 1: 1.3, 2: .1},
....: "X": dict(zip(range(3), np.random.randn(3)))
....: })
....:
In [46]: dft["id"] = dft.index
In [47]: dft
Out[47]:
A1970 A1980 B1970 B1980 X id
0 a d 2.5 3.2 -0.121306 0
1 b e 1.2 1.3 -0.097883 1
2 c f 0.7 0.1 0.695775 2
In [48]: pd.wide_to_long(dft, ["A", "B"], i="id", j="year")
Out[48]:
X A B
id year
0 1970 -0.121306 a 2.5
1 1970 -0.097883 b 1.2
2 1970 0.695775 c 0.7
0 1980 -0.121306 d 3.2
1 1980 -0.097883 e 1.3
2 1980 0.695775 f 0.1
Combination with statistics and GroupBy
You should be aware that combining pivot, stack, and ʻunstack` with GroupBy and basic Series and DataFrame statistical functions allows for very expressive and fast data manipulation.
In [49]: df
Out[49]:
exp A B A
animal cat dog cat dog
first second
bar one 0.895717 0.805244 -1.206412 2.565646
two 1.431256 1.340309 -1.170299 -0.226169
baz one 0.410835 0.813850 0.132003 -0.827317
two -0.076467 -1.187678 1.130127 -1.436737
foo one -1.413681 1.607920 1.024180 0.569605
two 0.875906 -2.211372 0.974466 -2.006747
qux one -0.410001 -0.078638 0.545952 -1.219217
two -1.226825 0.769804 -1.281247 -0.727707
In [50]: df.stack().mean(1).unstack()
Out[50]:
animal cat dog
first second
bar one -0.155347 1.685445
two 0.130479 0.557070
baz one 0.271419 -0.006733
two 0.526830 -1.312207
foo one -0.194750 1.088763
two 0.925186 -2.109060
qux one 0.067976 -0.648927
two -1.254036 0.021048
#Similar results in another way
In [51]: df.groupby(level=1, axis=1).mean()
Out[51]:
animal cat dog
first second
bar one -0.155347 1.685445
two 0.130479 0.557070
baz one 0.271419 -0.006733
two 0.526830 -1.312207
foo one -0.194750 1.088763
two 0.925186 -2.109060
qux one 0.067976 -0.648927
two -1.254036 0.021048
In [52]: df.stack().groupby(level=1).mean()
Out[52]:
exp A B
second
one 0.071448 0.455513
two -0.424186 -0.204486
In [53]: df.mean().unstack(0)
Out[53]:
exp A B
animal
cat 0.060843 0.018596
dog -0.413580 0.232430
Pivot table
pivot () has different data types (strings) , Numeric, etc.), but pandas provides pivot_table () for pivoting by aggregating numerical data. api / pandas.pivot_table.html # pandas.pivot_table) is also provided.
Spreadsheet-style pivot table using the function pivot_table () Can be created. See Cookbook for more advanced operations.
It takes some arguments.
--data: DataFrame object.
--value: A column or list of columns to aggregate.
--ʻIndex: Column, Grouper, array of the same length as the data, or a list of them. Keys to group by index in the pivot table. If an array is passed, it will be aggregated in the same way as the column values. --columns: An array of the same length as the columns, Grouper, or data, or a list of them. Keys to group by columns in the PivotTable. If an array is passed, it will be aggregated in the same way as the column values. --ʻAggfunc: Function used for aggregation. The default is numpy.mean.
Consider the following dataset.
In [54]: import datetime
In [55]: df = pd.DataFrame({'A': ['one', 'one', 'two', 'three'] * 6,
....: 'B': ['A', 'B', 'C'] * 8,
....: 'C': ['foo', 'foo', 'foo', 'bar', 'bar', 'bar'] * 4,
....: 'D': np.random.randn(24),
....: 'E': np.random.randn(24),
....: 'F': [datetime.datetime(2013, i, 1) for i in range(1, 13)]
....: + [datetime.datetime(2013, i, 15) for i in range(1, 13)]})
....:
In [56]: df
Out[56]:
A B C D E F
0 one A foo 0.341734 -0.317441 2013-01-01
1 one B foo 0.959726 -1.236269 2013-02-01
2 two C foo -1.110336 0.896171 2013-03-01
3 three A bar -0.619976 -0.487602 2013-04-01
4 one B bar 0.149748 -0.082240 2013-05-01
.. ... .. ... ... ... ...
19 three B foo 0.690579 -2.213588 2013-08-15
20 one C foo 0.995761 1.063327 2013-09-15
21 one A bar 2.396780 1.266143 2013-10-15
22 two B bar 0.014871 0.299368 2013-11-15
23 three C bar 3.357427 -0.863838 2013-12-15
[24 rows x 6 columns]
It's very easy to create a PivotTable from this data.
In [57]: pd.pivot_table(df, values='D', index=['A', 'B'], columns=['C'])
Out[57]:
C bar foo
A B
one A 1.120915 -0.514058
B -0.338421 0.002759
C -0.538846 0.699535
three A -1.181568 NaN
B NaN 0.433512
C 0.588783 NaN
two A NaN 1.000985
B 0.158248 NaN
C NaN 0.176180
In [58]: pd.pivot_table(df, values='D', index=['B'], columns=['A', 'C'], aggfunc=np.sum)
Out[58]:
A one three two
C bar foo bar foo bar foo
B
A 2.241830 -1.028115 -2.363137 NaN NaN 2.001971
B -0.676843 0.005518 NaN 0.867024 0.316495 NaN
C -1.077692 1.399070 1.177566 NaN NaN 0.352360
In [59]: pd.pivot_table(df, values=['D', 'E'], index=['B'], columns=['A', 'C'],
....: aggfunc=np.sum)
....:
Out[59]:
D ... E
A one three ... three two
C bar foo bar ... foo bar foo
B ...
A 2.241830 -1.028115 -2.363137 ... NaN NaN 0.128491
B -0.676843 0.005518 NaN ... -2.128743 -0.194294 NaN
C -1.077692 1.399070 1.177566 ... NaN NaN 0.872482
[3 rows x 12 columns]
The result object is a DataFrame that probably has a hierarchical index on the rows and columns. If no column name is specified for values, the PivotTable will add a hierarchy level to the column to contain all the data that can be aggregated.
In [60]: pd.pivot_table(df, index=['A', 'B'], columns=['C'])
Out[60]:
D E
C bar foo bar foo
A B
one A 1.120915 -0.514058 1.393057 -0.021605
B -0.338421 0.002759 0.684140 -0.551692
C -0.538846 0.699535 -0.988442 0.747859
three A -1.181568 NaN 0.961289 NaN
B NaN 0.433512 NaN -1.064372
C 0.588783 NaN -0.131830 NaN
two A NaN 1.000985 NaN 0.064245
B 0.158248 NaN -0.097147 NaN
C NaN 0.176180 NaN 0.436241
You can also use Grouper for the ʻindex and columnsarguments. For more information onGrouper`, see Grouping with Grouper (https://pandas.pydata.org/pandas-docs/stable/user_guide/groupby.html#groupby-specify).
In [61]: pd.pivot_table(df, values='D', index=pd.Grouper(freq='M', key='F'),
....: columns='C')
....:
Out[61]:
C bar foo
F
2013-01-31 NaN -0.514058
2013-02-28 NaN 0.002759
2013-03-31 NaN 0.176180
2013-04-30 -1.181568 NaN
2013-05-31 -0.338421 NaN
2013-06-30 -0.538846 NaN
2013-07-31 NaN 1.000985
2013-08-31 NaN 0.433512
2013-09-30 NaN 0.699535
2013-10-31 1.120915 NaN
2013-11-30 0.158248 NaN
2013-12-31 0.588783 NaN
You can beautifully render the output of a table with missing values omitted by calling to_string as needed.
In [62]: table = pd.pivot_table(df, index=['A', 'B'], columns=['C'])
In [63]: print(table.to_string(na_rep=''))
D E
C bar foo bar foo
A B
one A 1.120915 -0.514058 1.393057 -0.021605
B -0.338421 0.002759 0.684140 -0.551692
C -0.538846 0.699535 -0.988442 0.747859
three A -1.181568 0.961289
B 0.433512 -1.064372
C 0.588783 -0.131830
two A 1.000985 0.064245
B 0.158248 -0.097147
C 0.176180 0.436241
** pivot_table can also be used as an instance method of DataFrame. ** **
⇒ DataFrame.pivot_table()。
Add subtotal
Passing margins = True to pivot_table adds a special column and row called ʻAll` that aggregates the entire row and column category for each group.
Cross tabulation
Using crosstab (), you can use two (or more) You can cross-tabulate elements. If no array of values and aggregate function are passed, by default crosstab will calculate the frequency table of the elements.
It also receives the following arguments:
--ʻIndex: Array etc. Values to group by row. --columns: Array etc. Values to group by column. --values: Array etc. Optional. An array of values to aggregate according to the element. --ʻAggfunc: Function. Optional. If omitted, the frequency table is calculated.
--rownames: Sequence. The default is None. Must match the number of row arrays passed (the length of the array passed in the ʻindex argument). --colnames: Sequence. The default is None. Must match the number of column arrays passed (the length of the array passed in the columns argument). --margins: Boolean value. The default is False. Add subtotals to rows and columns. --normalizeBoolean value ・ {‘all’, ‘index’, ‘columns’} ・ {0,1}. The default isFalse`. Normalize by dividing all values by the sum of the values.
Unless a row or column name is specified in the crosstab, the name attribute of each passed Series is used.
For example
In [65]: foo, bar, dull, shiny, one, two = 'foo', 'bar', 'dull', 'shiny', 'one', 'two'
In [66]: a = np.array([foo, foo, bar, bar, foo, foo], dtype=object)
In [67]: b = np.array([one, one, two, one, two, one], dtype=object)
In [68]: c = np.array([dull, dull, shiny, dull, dull, shiny], dtype=object)
In [69]: pd.crosstab(a, [b, c], rownames=['a'], colnames=['b', 'c'])
Out[69]:
b one two
c dull shiny dull shiny
a
bar 1 0 0 1
foo 2 1 1 0
If crosstab receives only two Series, a frequency table is returned.
In [70]: df = pd.DataFrame({'A': [1, 2, 2, 2, 2], 'B': [3, 3, 4, 4, 4],
....: 'C': [1, 1, np.nan, 1, 1]})
....:
In [71]: df
Out[71]:
A B C
0 1 3 1.0
1 2 3 1.0
2 2 4 NaN
3 2 4 1.0
4 2 4 1.0
In [72]: pd.crosstab(df['A'], df['B'])
Out[72]:
B 3 4
A
1 1 0
2 1 3
If the data passed has Categorical data, then the ** all ** categories will be included in the crosstab, even if the actual data does not contain instances of a particular category.
In [73]: foo = pd.Categorical(['a', 'b'], categories=['a', 'b', 'c'])
In [74]: bar = pd.Categorical(['d', 'e'], categories=['d', 'e', 'f'])
In [75]: pd.crosstab(foo, bar)
Out[75]:
col_0 d e
row_0
a 1 0
b 0 1
Normalization
The frequency table can also be normalized to display percentages instead of counts using the normalize argument.
In [76]: pd.crosstab(df['A'], df['B'], normalize=True)
Out[76]:
B 3 4
A
1 0.2 0.0
2 0.2 0.6
normalize can also be normalized for each row or column.
In [77]: pd.crosstab(df['A'], df['B'], normalize='columns')
Out[77]:
B 3 4
A
1 0.5 0.0
2 0.5 1.0
If you pass the third Series and the aggregate function (ʻaggfunc) to crosstab, the function for the value in the third Series of each group defined in the first two Series`s. Is applied.
In [78]: pd.crosstab(df['A'], df['B'], values=df['C'], aggfunc=np.sum)
Out[78]:
B 3 4
A
1 1.0 NaN
2 1.0 2.0
Add subtotal
Finally, you can add subtotals and normalize their output.
In [79]: pd.crosstab(df['A'], df['B'], values=df['C'], aggfunc=np.sum, normalize=True,
....: margins=True)
....:
Out[79]:
B 3 4 All
A
1 0.25 0.0 0.25
2 0.25 0.5 0.75
All 0.50 0.5 1.00
Binning
The cut () function performs grouping calculations for input array values. I will do it. It is often used to convert continuous variables to discrete or categorical variables.
In [80]: ages = np.array([10, 15, 13, 12, 23, 25, 28, 59, 60])
In [81]: pd.cut(ages, bins=3)
Out[81]:
[(9.95, 26.667], (9.95, 26.667], (9.95, 26.667], (9.95, 26.667], (9.95, 26.667], (9.95, 26.667], (26.667, 43.333], (43.333, 60.0], (43.333, 60.0]]
Categories (3, interval[float64]): [(9.95, 26.667] < (26.667, 43.333] < (43.333, 60.0]]
Passing an integer as the bins argument will form a monospaced bin. You can also specify a custom bin edge.
In [82]: c = pd.cut(ages, bins=[0, 18, 35, 70])
In [83]: c
Out[83]:
[(0, 18], (0, 18], (0, 18], (0, 18], (18, 35], (18, 35], (18, 35], (35, 70], (35, 70]]
Categories (3, interval[int64]): [(0, 18] < (18, 35] < (35, 70]]
If you pass ʻIntervalIndex to the bins` argument, it will be used to bin the data.
pd.cut([25, 20, 50], bins=c.categories)
Calculation of indicator variables and dummy variables
Using get_dummies () makes categorical variables "dummy" and "markers" Can be converted to DataFrame. For example, from a DataFrame column (Series) with k different values, you can create a DataFrame containing k columns consisting of 1s and 0s.
In [84]: df = pd.DataFrame({'key': list('bbacab'), 'data1': range(6)})
In [85]: pd.get_dummies(df['key'])
Out[85]:
a b c
0 0 1 0
1 0 1 0
2 1 0 0
3 0 0 1
4 1 0 0
5 0 1 0
Prefixing column names is useful, for example, when merging the result with the original DataFrame.
In [86]: dummies = pd.get_dummies(df['key'], prefix='key')
In [87]: dummies
Out[87]:
key_a key_b key_c
0 0 1 0
1 0 1 0
2 1 0 0
3 0 0 1
4 1 0 0
5 0 1 0
In [88]: df[['data1']].join(dummies)
Out[88]:
data1 key_a key_b key_c
0 0 0 1 0
1 1 0 1 0
2 2 1 0 0
3 3 0 0 1
4 4 1 0 0
5 5 0 1 0
This function is often used with discretized functions like cut.
In [89]: values = np.random.randn(10)
In [90]: values
Out[90]:
array([ 0.4082, -1.0481, -0.0257, -0.9884, 0.0941, 1.2627, 1.29 ,
0.0824, -0.0558, 0.5366])
In [91]: bins = [0, 0.2, 0.4, 0.6, 0.8, 1]
In [92]: pd.get_dummies(pd.cut(values, bins))
Out[92]:
(0.0, 0.2] (0.2, 0.4] (0.4, 0.6] (0.6, 0.8] (0.8, 1.0]
0 0 0 1 0 0
1 0 0 0 0 0
2 0 0 0 0 0
3 0 0 0 0 0
4 1 0 0 0 0
5 0 0 0 0 0
6 0 0 0 0 0
7 1 0 0 0 0
8 0 0 0 0 0
9 0 0 1 0 0
See also Series.str.get_dummies Please give me.
get_dummies () can also receive DataFrame. By default, all categorical variables (in the statistical sense, that is, those with * object * or * categorical * data types) are encoded into dummy variables.
In [93]: df = pd.DataFrame({'A': ['a', 'b', 'a'], 'B': ['c', 'c', 'b'],
....: 'C': [1, 2, 3]})
....:
In [94]: pd.get_dummies(df)
Out[94]:
C A_a A_b B_b B_c
0 1 1 0 0 1
1 2 0 1 0 1
2 3 1 0 1 0
All non-object columns are included in the output as is. You can control which columns are encoded with the columns argument.
In [95]: pd.get_dummies(df, columns=['A'])
Out[95]:
B C A_a A_b
0 c 1 1 0
1 c 2 0 1
2 b 3 1 0
You can see that the B column is still included in the output, but it is not encoded. If you don't want to include it in the output, drop B before calling get_dummies.
As with the Series version, you can pass the values for prefix and prefix_sep. By default, the column name is used as the prefix and "_" is used as the column name delimiter. prefix and prefix_sep can be specified in the following three ways.
--String: Use the same value for the prefix or prefix_sep of each column to encode.
--List: Must be as long as the number of columns to be encoded.
--Dictionary: Map column names to prefixes.
In [96]: simple = pd.get_dummies(df, prefix='new_prefix')
In [97]: simple
Out[97]:
C new_prefix_a new_prefix_b new_prefix_b new_prefix_c
0 1 1 0 0 1
1 2 0 1 0 1
2 3 1 0 1 0
In [98]: from_list = pd.get_dummies(df, prefix=['from_A', 'from_B'])
In [99]: from_list
Out[99]:
C from_A_a from_A_b from_B_b from_B_c
0 1 1 0 0 1
1 2 0 1 0 1
2 3 1 0 1 0
In [100]: from_dict = pd.get_dummies(df, prefix={'B': 'from_B', 'A': 'from_A'})
In [101]: from_dict
Out[101]:
C from_A_a from_A_b from_B_b from_B_c
0 1 1 0 0 1
1 2 0 1 0 1
2 3 1 0 1 0
To avoid collinearity when using the results in a statistical model, it may be useful to keep only the k-1 level of the categorical variable. You can switch to this mode using drop_first.
In [102]: s = pd.Series(list('abcaa'))
In [103]: pd.get_dummies(s)
Out[103]:
a b c
0 1 0 0
1 0 1 0
2 0 0 1
3 1 0 0
4 1 0 0
In [104]: pd.get_dummies(s, drop_first=True)
Out[104]:
b c
0 0 0
1 1 0
2 0 1
3 0 0
4 0 0
If the column contains only one level, it will be omitted in the result.
In [105]: df = pd.DataFrame({'A': list('aaaaa'), 'B': list('ababc')})
In [106]: pd.get_dummies(df)
Out[106]:
A_a B_a B_b B_c
0 1 1 0 0
1 1 0 1 0
2 1 1 0 0
3 1 0 1 0
4 1 0 0 1
In [107]: pd.get_dummies(df, drop_first=True)
Out[107]:
B_b B_c
0 0 0
1 1 0
2 0 0
3 1 0
4 0 1
By default, the new column will be np.uint8dtype. Use the dtype argument to select a different data type.
In [108]: df = pd.DataFrame({'A': list('abc'), 'B': [1.1, 2.2, 3.3]})
In [109]: pd.get_dummies(df, dtype=bool).dtypes
Out[109]:
B float64
A_a bool
A_b bool
A_c bool
dtype: object
_ From version 0.23.0 _
Element value (label encoding)
To encode a one-dimensional value as an enum, factorize ()Use the.
In [110]: x = pd.Series(['A', 'A', np.nan, 'B', 3.14, np.inf])
In [111]: x
Out[111]:
0 A
1 A
2 NaN
3 B
4 3.14
5 inf
dtype: object
In [112]: labels, uniques = pd.factorize(x)
In [113]: labels
Out[113]: array([ 0, 0, -1, 1, 2, 3])
In [114]: uniques
Out[114]: Index(['A', 'B', 3.14, inf], dtype='object')
Note that factorize is similar to numpy.unique, but handles NaN differently.
: ballot_box_with_check: ** Note ** The following
numpy.uniquefails withTypeErrorin Python 3 due to an alignment bug. For more information, please see here.
In [1]: x = pd.Series(['A', 'A', np.nan, 'B', 3.14, np.inf])
In [2]: pd.factorize(x, sort=True)
Out[2]:
(array([ 2, 2, -1, 3, 0, 1]),
Index([3.14, inf, 'A', 'B'], dtype='object'))
In [3]: np.unique(x, return_inverse=True)[::-1]
Out[3]: (array([3, 3, 0, 4, 1, 2]), array([nan, 3.14, inf, 'A', 'B'], dtype=object))
: ballot_box_with_check: ** Note ** If you want to treat a column as a categorical variable (like R factor),
df ["cat_col"] = pd.Categorical (df ["col"])ordf ["cat_col"] = You can use df ["col"] .astype ("category"). For complete documentation onCategorical, see Introduction to Categorical //pandas.pydata.org/pandas-docs/stable/user_guide/categorical.html#categorical) and API Documentation See # api-arrays-categorical).
Example
This section contains frequently asked questions and examples. The column names and associated column values have names that correspond to how this DataFrame is pivoted in the answers below.
In [115]: np.random.seed([3, 1415])
In [116]: n = 20
In [117]: cols = np.array(['key', 'row', 'item', 'col'])
In [118]: df = cols + pd.DataFrame((np.random.randint(5, size=(n, 4))
.....: // [2, 1, 2, 1]).astype(str))
.....:
In [119]: df.columns = cols
In [120]: df = df.join(pd.DataFrame(np.random.rand(n, 2).round(2)).add_prefix('val'))
In [121]: df
Out[121]:
key row item col val0 val1
0 key0 row3 item1 col3 0.81 0.04
1 key1 row2 item1 col2 0.44 0.07
2 key1 row0 item1 col0 0.77 0.01
3 key0 row4 item0 col2 0.15 0.59
4 key1 row0 item2 col1 0.81 0.64
.. ... ... ... ... ... ...
15 key0 row3 item1 col1 0.31 0.23
16 key0 row0 item2 col3 0.86 0.01
17 key0 row4 item0 col3 0.64 0.21
18 key2 row2 item2 col0 0.13 0.45
19 key0 row2 item0 col4 0.37 0.70
[20 rows x 6 columns]
Pivot with a single aggregate
Suppose you want to pivot df so that the value of col is the column, the value of row is the index, and the average of val0 is the table value. At this time, the resulting DataFrame looks like this:
col col0 col1 col2 col3 col4
row
row0 0.77 0.605 NaN 0.860 0.65
row2 0.13 NaN 0.395 0.500 0.25
row3 NaN 0.310 NaN 0.545 NaN
row4 NaN 0.100 0.395 0.760 0.24
To find this, use pivot_table (). Note that ʻaggfunc ='mean'` is the default behavior, although explicitly mentioned here.
In [122]: df.pivot_table(
.....: values='val0', index='row', columns='col', aggfunc='mean')
.....:
Out[122]:
col col0 col1 col2 col3 col4
row
row0 0.77 0.605 NaN 0.860 0.65
row2 0.13 NaN 0.395 0.500 0.25
row3 NaN 0.310 NaN 0.545 NaN
row4 NaN 0.100 0.395 0.760 0.24
You can also use the fill_value parameter to replace missing values.
In [123]: df.pivot_table(
.....: values='val0', index='row', columns='col', aggfunc='mean', fill_value=0)
.....:
Out[123]:
col col0 col1 col2 col3 col4
row
row0 0.77 0.605 0.000 0.860 0.65
row2 0.13 0.000 0.395 0.500 0.25
row3 0.00 0.310 0.000 0.545 0.00
row4 0.00 0.100 0.395 0.760 0.24
Also, be aware that you can pass other aggregate functions. For example, you can pass sum.
In [124]: df.pivot_table(
.....: values='val0', index='row', columns='col', aggfunc='sum', fill_value=0)
.....:
Out[124]:
col col0 col1 col2 col3 col4
row
row0 0.77 1.21 0.00 0.86 0.65
row2 0.13 0.00 0.79 0.50 0.50
row3 0.00 0.31 0.00 1.09 0.00
row4 0.00 0.10 0.79 1.52 0.24
Another aggregation is to calculate how often columns and rows occur at the same time (called "cross aggregation"). To do this, pass size to the ʻaggfunc` parameter.
In [125]: df.pivot_table(index='row', columns='col', fill_value=0, aggfunc='size')
Out[125]:
col col0 col1 col2 col3 col4
row
row0 1 2 0 1 1
row2 1 0 2 1 2
row3 0 1 0 2 0
row4 0 1 2 2 1
Pivot with multiple aggregates
You can also perform multiple aggregations. For example, you can pass a list to the ʻaggfunc argument to perform both the sum and the mean mean.
In [126]: df.pivot_table(
.....: values='val0', index='row', columns='col', aggfunc=['mean', 'sum'])
.....:
Out[126]:
mean sum
col col0 col1 col2 col3 col4 col0 col1 col2 col3 col4
row
row0 0.77 0.605 NaN 0.860 0.65 0.77 1.21 NaN 0.86 0.65
row2 0.13 NaN 0.395 0.500 0.25 0.13 NaN 0.79 0.50 0.50
row3 NaN 0.310 NaN 0.545 NaN NaN 0.31 NaN 1.09 NaN
row4 NaN 0.100 0.395 0.760 0.24 NaN 0.10 0.79 1.52 0.24
You can pass a list to the values parameter to aggregate across multiple value columns.
In [127]: df.pivot_table(
.....: values=['val0', 'val1'], index='row', columns='col', aggfunc=['mean'])
.....:
Out[127]:
mean
val0 val1
col col0 col1 col2 col3 col4 col0 col1 col2 col3 col4
row
row0 0.77 0.605 NaN 0.860 0.65 0.01 0.745 NaN 0.010 0.02
row2 0.13 NaN 0.395 0.500 0.25 0.45 NaN 0.34 0.440 0.79
row3 NaN 0.310 NaN 0.545 NaN NaN 0.230 NaN 0.075 NaN
row4 NaN 0.100 0.395 0.760 0.24 NaN 0.070 0.42 0.300 0.46
You can pass a list as a column parameter to subdivide into multiple columns.
In [128]: df.pivot_table(
.....: values=['val0'], index='row', columns=['item', 'col'], aggfunc=['mean'])
.....:
Out[128]:
mean
val0
item item0 item1 item2
col col2 col3 col4 col0 col1 col2 col3 col4 col0 col1 col3 col4
row
row0 NaN NaN NaN 0.77 NaN NaN NaN NaN NaN 0.605 0.86 0.65
row2 0.35 NaN 0.37 NaN NaN 0.44 NaN NaN 0.13 NaN 0.50 0.13
row3 NaN NaN NaN NaN 0.31 NaN 0.81 NaN NaN NaN 0.28 NaN
row4 0.15 0.64 NaN NaN 0.10 0.64 0.88 0.24 NaN NaN NaN NaN
Expand the columns of the list
_ From version 0.25.0 _
The column values may look like a list.
In [129]: keys = ['panda1', 'panda2', 'panda3']
In [130]: values = [['eats', 'shoots'], ['shoots', 'leaves'], ['eats', 'leaves']]
In [131]: df = pd.DataFrame({'keys': keys, 'values': values})
In [132]: df
Out[132]:
keys values
0 panda1 [eats, shoots]
1 panda2 [shoots, leaves]
2 panda3 [eats, leaves]
Using [ʻexplode () ](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.explode.html#pandas.Series.explode), values` You can "explode" columns and convert each list element into a separate row. This duplicates the index value from the original row.
In [133]: df['values'].explode()
Out[133]:
0 eats
0 shoots
1 shoots
1 leaves
2 eats
2 leaves
Name: values, dtype: object
You can also expand the columns of DataFrame.
In [134]: df.explode('values')
Out[134]:
keys values
0 panda1 eats
0 panda1 shoots
1 panda2 shoots
1 panda2 leaves
2 panda3 eats
2 panda3 leaves
Series.explode () is an empty list Replace with np.nan and keep the scalar entry. The resulting Series data type is always ʻobject`.
In [135]: s = pd.Series([[1, 2, 3], 'foo', [], ['a', 'b']])
In [136]: s
Out[136]:
0 [1, 2, 3]
1 foo
2 []
3 [a, b]
dtype: object
In [137]: s.explode()
Out[137]:
0 1
0 2
0 3
1 foo
2 NaN
3 a
3 b
dtype: object
As a typical example, suppose a column has a comma-separated string and you want to expand it.
In [138]: df = pd.DataFrame([{'var1': 'a,b,c', 'var2': 1},
.....: {'var1': 'd,e,f', 'var2': 2}])
.....:
In [139]: df
Out[139]:
var1 var2
0 a,b,c 1
1 d,e,f 2
You could easily create a vertical DataFrame by performing expansion and chain operations.
In [140]: df.assign(var1=df.var1.str.split(',')).explode('var1')
Out[140]:
var1 var2
0 a 1
0 b 1
0 c 1
1 d 2
1 e 2
1 f 2
Recommended Posts