Get rid of DICOM images in Python
To put it simply, DICOM images are images that are handled on the communication protocol (DICOM) for exchanging medical images between devices, and various additional information (patient information, patient information, etc.) to the images. It is the type of device, the information of the image itself, and various metadata).
When a doctor performs an examination such as CT or MRI, some hospitals may ask you to use the image as data. If the format at this time is the extension .dcm, it is a DICOM image.
Preparing to work with DICOM in Python
If you want to work with DICOM images in Python, it seems that you will use pydicom.
% anaconda show conda-forge/pydicom
Using Anaconda API: https://api.anaconda.org
Name: pydicom
Summary: Pure python package for DICOM medical file reading and writing
Access: public
Package Types: conda
Versions:
+ 0.9.9
To install this package with conda run:
conda install --channel https://conda.anaconda.org/conda-forge pydicom
That's why `conda install -c https://conda.anaconda.org/conda-forge pydicom`.
Prepare a sample DICOM image
If you unfortunately don't have your own MRI image, download a sample image. There is DICOM Image Library on the Osirix site, which is famous as a DICOM Viewer, so get a suitable one from here. For the time being, I downloaded an MRI image of a brain tumor called BRAINIX. If you unzip this .zip file, you will see images (T1 weighted, etc.) that have been processed in various ways for the interpretation doctor to see after imaging with MRI. Among them, "T1-3D-FFE-C" I decided to use 100 files in the folder "-\ 801 /".
Read DICOM images
Below is an example of how to load an image using the pydicom function.
Read one DICOM image and refer to the metadata
Load the image and display the DICOM header.
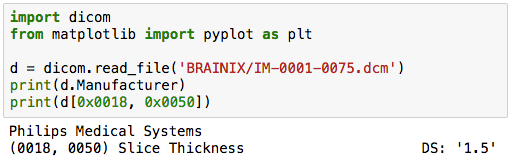
import dicom
d = dicom.read_file('BRAINIX/IM-0001-0075.dcm')
print(d)
By the way, if you want to display specific metadata, do as follows.
print(d.Manufacturer)
Apparently, the device manufacturer is "Philips Medical Systems".
print(d[0x0018, 0x0050])
Apparently, the Slice thickness is 1.5mm.
In this way, DICOM metadata has a key name called a tag, and the keys are numbered. And with pydicom, you can display either the key name or the number.
Read one DICOM image and display the image
Since the substance of the image part of the DICOM image can be extracted in the array format of numpy, it can be displayed by using either opencv or matplotlib.

Reads multiple DICOM images and converts them into one continuous tomographic image
By the way, the BRAINIX image used this time is a series of 100 images with a thickness of 1.5 mm. These 100 images can be read and imported into one 3D array.
import dicom
from matplotlib import pyplot as plt
%matplotlib inline
root_dir = './BRAINIX'
dcms = []
for d, s, fl in os.walk(root_dir):
for fn in fl:
if ".dcm" in fn.lower():
dcms.append(os.path.join(d, fn))
ref_dicom = dicom.read_file(dcms[0])
d_array = np.zeros((ref_dicom.Rows, ref_dicom.Columns, len(dcms)), dtype=ref_dicom.pixel_array.dtype)
for dcm in dcms:
d = dicom.read_file(dcm)
d_array[:, :, dcms.index(dcm)] = d.pixel_array
In short, all the files under the BRAINIX folder with the extension .dcm are read, and the contents are copied in order to a three-dimensional array called d_array.
If you want to display any slice, for example the 50th slice, just do `pyplt.imshow (d_array [:, :, 49])`.
Now that it has been read into a 3D array, it is possible to display 3 cross sections.
import os
import dicom
from matplotlib import pyplot as plt
%matplotlib inline
#Read the dcm file under the BRAINIX folder
root_dir = './BRAINIX'
dcms = []
for d, s, fl in os.walk(root_dir):
for fn in fl:
if ".dcm" in fn.lower():
dcms.append(os.path.join(d, fn))
ref_dicom = dicom.read_file(dcms[0])
d_array = np.zeros((ref_dicom.Rows, ref_dicom.Columns, len(dcms)), dtype=ref_dicom.pixel_array.dtype)
for dcm in dcms:
d = dicom.read_file(dcm)
d_array[:, :, dcms.index(dcm)] = d.pixel_array
#3 Cross section display
print(d_array.shape)
plt.subplot(1, 3, 1)
plt.imshow(d_array[127, :, :])
plt.subplot(1, 3, 2)
plt.imshow(d_array[:, 127, :])
plt.subplot(1, 3, 3)
plt.imshow(d_array[:, :, 49])

Process and save DICOM images
If you want to process the read DICOM image and save it by overwriting the original DICOM image, you need to edit the array of pixel_array.
However, the original image does not change just by editing the array of pixel_array. This is because the actual image data of the DICOM image is stored in an array called PixelData, and the interface for reading it is pixel_array. Therefore, if the contents of the edited pixel_array are saved in an array called img, for example, it is necessary to write back the contents to Pixel Data.
d = dicom.read_file(dcm)
img = d.pixel_array
#Processing of img
d.PixelData = img.tostring()
d.save_as(dcm)
Try cropping a DICOM image into a circle
I don't know what it is, but I'll try the work of "cutting out the DICOM image only in the center part with a circle with a diameter half the length and width of the image".
With the following code, an image with a diameter of half the length and width of the image is 1 inside and 0 outside. If the radius is r, the vertical and horizontal size of the image can be obtained from ref_dicom.pixel_array.shape, so set 1/4 of that size.
r = ref_dicom.pixel_array.shape[0] / 4
x, y = np.indices((ref_dicom.Rows, ref_dicom.Columns))
circle = (x - (ref_dicom.Columns / 2))**2 + (y - (ref_dicom.Rows / 2))**2 < r**2
mask = circle.astype(int)
Multiplying this mask, the inside of the circle is 1 and the outside is 0, so you can cut out a circle.
###Read multiple DICOM image files and cut out the center part in a circle
import os
import dicom
from matplotlib import pyplot as plt
%matplotlib inline
#Read the dcm file under the BRAINIX folder
root_dir = './BRAINIX'
dcms = []
for d, s, fl in os.walk(root_dir):
for fn in fl:
if ".dcm" in fn.lower():
dcms.append(os.path.join(d, fn))
ref_dicom = dicom.read_file(dcms[0])
#Create a mask to cut out into a circle
r = ref_dicom.pixel_array.shape[0] / 4
x, y = np.indices((ref_dicom.Rows, ref_dicom.Columns))
circle = (x - (ref_dicom.Columns / 2))**2 + (y - (ref_dicom.Rows / 2))**2 < r**2
mask = circle.astype(int)
#Perform mask processing on all images
d_array = np.zeros((ref_dicom.Rows, ref_dicom.Columns, len(dcms)), dtype=ref_dicom.pixel_array.dtype)
for dcm in dcms:
d = dicom.read_file(dcm)
img = d.pixel_array * mask
d_array[:, :, dcms.index(dcm)] = img
#3 Cross section display
plt.figure(figsize=(8, 6))
plt.subplot(1, 3, 1)
plt.imshow(d_array[127, :, :])
plt.subplot(1, 3, 2)
plt.imshow(d_array[:, 127, :])
plt.subplot(1, 3, 3)
plt.imshow(d_array[:, :, 49])
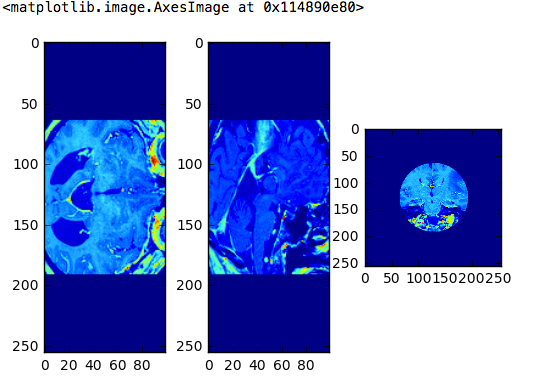
When dealing with DICOM images in Python, it looks like this.
Today's code
- https://github.com/fukuit/Python_SelfLearning/tree/master/dicom
Recommended Posts